York, 7th –9th of February 2024 MRC National Mouse Genetics Network Meeting Report
The Network was welcomed by the stunning city of York for the largest meeting we have had since our launch in April 2022.
The meeting was organised with three main purposes:
- Provide a space for our Early Career Researchers (ECRs) to come together and discuss topics that are key to their current work and career development.
- Discuss scientific progress and plans, including for the newly launched Ageing Cluster.
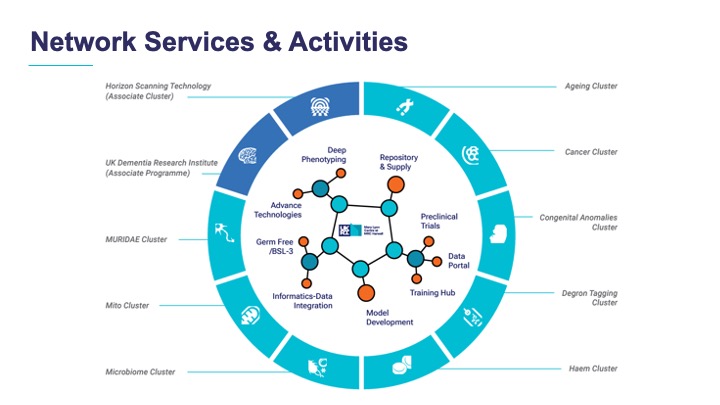
- Gather the Network’s Associate Members to introduce the new cluster and outline what the Network can provide for them, including being a source of funding.
The meeting was therefore spread over three days, each one focused on one of those aims.
Focus on the Network’s Early Career Researchers
The first part of the meeting was held at the beautiful Mansion House in the centre of town and was organised and run by the Network’s ECRs. Sixty delegates attended a vibrant afternoon of talks and debate. An introduction to the Network by Sara Wells, Director of the Mary Lyon Centre at MRC Harwell (MLC), the Hub of the Network, was followed by a series of three invited speakers who provided valuable insights into their career paths and the elements that made it possible for them to reach their current positions.



The second part of the afternoon was dedicated to a debate focused on animal experimentation in which the motion was ”This house believes that mouse models will be used in 50 years’ time.”
The debate was split into four subsections to dissect the key elements of animal modelling for medical research: disease modelling, disease treatment, future technologies, and ethics. Arguments for and against the motion were put forward by the participants and a final vote was held, in which the audience agreed that mouse models would still be used.
Aside from the result of the vote, all arguments were eloquently developed and offered food for thought to everyone present.
The networking opportunity presented by this event is a key element of the work of the Network in fostering and supporting interactions within and between clusters at all levels.
The new Ageing Cluster and the science so far
Day 2 of the meeting was dedicated to Network members and scientific progress discussions as we moved to the National STEM Learning Centre, but the main event of the day was the official introduction of the new Ageing Cluster. The two cluster leads, Laura Greaves and Walid Khaled, outlined their ambitious proposal and the composition of their team, also highlighting the numerous points of contact with the existing clusters.

ECRs were then given the chance to talk about their work in a series of four flash talks.
Eoghan Mulholland, from the Cancer Cluster, spoke about stromal remodelling and tissue co-evolution in intestinal cancers. Sevda Boyanova, a researcher in Frances Wiseman’s group at the UK Dementia Research Institute (DRI), reported on novel cognitive and sensory phenotyping in two new mouse models of ALS/FTD (amyotrophic lateral sclerosis/frontotemporal dementia). Alasdair Duguid, from the Haem Cluster, discussed immunotherapy failures in central nervous system (CNS) leukaemia. Amelia Bryers, a PhD student in the MURIDAE Cluster, reported on serotonin system function in a mouse model of neurodevelopmental disorder. Delegates were able to ask questions during a poster session after the talks.
After a break and some networking, Carlo Viscomi of the University of Padua (Italy) reported on adenovirus-mediated gene therapy trials for mitochondrial diseases in mouse models highlighting their specificity and efficiency.

Claus Nerlov of the Haem Cluster gave a detailed update of his work on the role of Kit ligand in hematopoietic niches. This was followed by Jen Morton of the Cancer Cluster who spoke about modelling microenvironmental changes in mouse models of pancreatic cancer with the view to improving therapy outcomes.
It was then the turn of the first keynote speaker of the day. Elizabeth (Lizzy) Fisher, from University College London, gave a fascinating insight into the world of humanised mouse models, specifically in neurodegenerative disease. Lizzy dissected the reasons behind the use of such models, outlining the current state of research in this area and ways in which the incredible potential of these mouse mutants can be maximised, not shying away from the technical, financial and ethical challenges associated with their use.
The afternoon sessions were kicked off by Stephen Twigg of the Congenital Anomalies Cluster who reported on some of the work carried out by the team on rare congenital syndromes that collectively affect 6–8% of the population. Stephen highlighted how modern sequencing technologies are leading to advances in genetic diagnosis, which has a profound impact on affected individuals and families, and allows for the prompt study of variants of uncertain significance in animal models; such variants can be used to more easily confirm diagnoses and can indicate therapeutic avenues.
Ben Davies of The Francis Crick Institute and lead of the Horizon Scanning Associate Cluster followed Lizzy’s keynote presentation with a technical insight into the technologies available for the generation of complex mouse mutants with large insertions, such as the humanised models previously described.
Anthony Isles of the MURIDAE Cluster described in some detail some of the work of the team on a gene strongly associated with schizophrenia and developmental disorders, and how the relevant mouse models can support the identification of the molecular pathways leading to the phenotype, including the involvement of placental programming.
Frances Wiseman of the UK DRI and lead of the Network’s DRI Associate Programme spoke to us about the current understanding of neuroimmune biology in Down syndrome-associated Alzheimer’s disease obtained using mouse models.
ECR flash talks followed, with Pedro Silva-Pinheiro, of the Mitochondria Cluster, talking about a library of base editors for the precise ablation of genes in the mitochondrial genome. Lorraine Eley, from the Congenital Anomalies Cluster, showed the creation of a mouse–human heart atlas for the comparison of normal and abnormal phenotypes, made possible by the creation and accurate phenotyping of mouse models of congenital heart disease and the annotation of human foetal heart specimens. Xiang Li, from The Francis Crick Institute, explored the role of integrases for efficient gene modification in addition to the BAC recombinase-mediated cassette exchange (RMCE) method previously described by Ben Davies, to create a more comprehensive toolkit fit for the requirements of complex model generation. Henry Taylor, of the Microbiome Cluster, described how some gain-of-function mutations in the SYK gene can alter the host immune response to intestinal flora by causing early-onset autoinflammatory disease and a B-cell defect linked to increased susceptibility to intestinal pathogens.
The last session of the day began with Roland Wolf of the Degron Tagging Cluster describing a humanised cytochrome P450 mouse model that has a significant advantage over wildtype mice for drug development as a result of its closer alignment with human drug metabolism, which will allow faster and more relevant data to be collected https://pubmed.ncbi.nlm.nih.gov/30910785/, https://www.pnas.org/doi/10.1073/pnas.2315069121
Rob Finn, from the Microbiome Cluster, concluded the cluster science updates by reporting results on computational analysis aimed at mapping the diversity of the mouse microbiome using available databases, highlighting the numerous applications available to the community and the way in which the data will be accessible.
The final keynote presentation and last talk of a very busy day was given by Richard Coward of the University of Bristol, who described how transgenic mice were instrumental in defining the mechanism of action of a bacterial toxin in the leading cause of global kidney failure and acute kidney injury in children. His work using carefully engineered mouse models (also reported here) will have a direct impact on diagnosis and therapeutic interventions for this devastating condition.

A panel discussion on the key issues of preclinical in vivo modelling concluded the day.
A day for the Network’s Associates
The final day of the meeting was dedicated to the Network’s Associates to highlight how they can interact with the Network and benefit from access to resources including funding.
After an introduction by the Network’s Director Owen Sansom, Lydia Teboul from the MLC gave everyone a flavour of how her team works towards introducing complex mutations into the mouse genome, describing how each allele is extensively quality-controlled and validated using a combination of techniques that allow for the most accurate analysis in the fastest possible time, and with constant attention to the value-for-money element of the equation.
Ilaria Cervellini and Sonia Bains, also both from the MLC, provided delegates with a comprehensive overview of the phenotyping services on offer at the centre, with attention to new technologies and the availability of the home-cage monitoring system developed in the last ten years, which is now regularly used to produce new exciting data about mouse behaviour in an undisturbed environment.
The following session began with the launch of the second round of the Network’s Director’s Fund, a call for pilot/proof of principle projects spanning 6–18 months and with awards ranging between £20,000 and £100,000. The projects must be aligned to the Network’s activities and the lead applicants should be Network Associates or ECRs.
The announcement was followed by two examples of projects funded in the previous call. William Grey from the University of York introduced the UK Patient-Derived Xenograft Biobank project run in collaboration with Dominique Bonnet of The Francis Crick Institute and Sara Wells, Director of the MLC, as a platform for translational cancer research. Jethro Johnson from the Microbiome Cluster presented his project which is focused on looking at the extent to which mouse microbiomes vary between facilities across the Network, producing some stark evidence about how the influence of a facility has a greater effect than that of the host’s genetic background.
The next session was opened by the introduction of the new Ageing Cluster to the Associates by Laura Greaves. This was followed by Stephen Twigg of the Congenital Anomalies Cluster promoting the new Variant of Uncertain Significance submission portal, which is live on the Network’s website and will allow clinicians and human geneticists to make use of the cluster’s resources and expertise to validate newly identified and clinically relevant variants in human patients.

A great example of a cross-cluster, multidisciplinary approach project was presented by Johan Vande Voorde of the University of Glasgow and CRUK Scotland Institute, who spoke about the effect of gut microbiota on colorectal cancers as demonstrated by studies using germ-free and gnotobiotic animals. He was followed by Rory Steven of the National Physical Laboratory, who provided examples of how multimodal mass spectrometry imaging (MSI) can be used to resolve imaging limitations from other systems and to achieve great resolution at the organ, cellular and molecular levels to study disease, drug metabolism and to answer key biological questions.
Frances Wiseman introduced the UK DRI Animal Model Programme, which is an associated element of the Network, and highlighted its aims and the tools available to the community via the programme as well as through the MLC.
David Walters of Cancer Research Horizons gave an introduction to the Functional Genomics Centre and outlined its vision to become a world-leading centre of excellence in genetic screening, cancer models, CRISPR vector design and computational approaches to big data. Its goal is to identify and validate novel drug targets and to understand resistance mechanisms in the search for new cancer therapeutics.
David Kent, from the Haem Cluster, introduced the Network’s training opportunities, which are available to the associates, including the Training Working Group, highlighting the dense programme of courses and events already in place. David also spoke about the newly developed Network’s doctoral training programme to be launched in the Autumn, emphasising its key ambition to fill a current skills gap in the field of in vivo model genetics and to train the future generation of preclinical researchers in the area.
Finally, Rob Pitceathly of the MitoCluster presented all the Network’s current working groups, highlighting their objectives and reasons why the associates should join these to get involved in discussions and activities promoted by the Network, including the upcoming ‘Mouse Models of Mitochondrial Disease: Theory and Practice’ training course to be held at the Advance Training Centre at MRC Harwell.
It was a packed programme that left all delegates full of new ideas and plans for future interactions and collaborations.