Announcing the first set of projects supported by the Director’s Fund

We are pleased to announce the first set of projects to receive funding from the MRC National Mouse Genetics Network Director’s Fund, which include projects relating to the microbiome, mitochondria, cholesterol metabolism, cancer, establishment of a patient-derived xenograft biobank, and development of a tool for the degradation of target proteins. More information about all the funded projects can be found on the Projects page.
The Director’s Fund is an annual award for a total of £200,000 in the first year and £100,000 for the following years of the Network’s lifespan. Individual awards receive funding of between £20,000 and £100,000 for projects typically running for periods of 6–18 months. The Fund aims to support projects that are aligned in some way with the Network’s ongoing activities, such as through collaborations between existing clusters, projects that relate to the work of a specific cluster, or projects that develop tools, technologies, and resources of interest to the Network or specific clusters.
One project that has a clear link to a particular cluster is the project Assessing gut microbiome heterogeneity across the National Mouse Genetics Network, although it will also make use of the involvement of a spread of facilities involved in the Network. Improving experimental reproducibility is a major goal in mouse genetics research. It is well established that the microbiomes of mice vary between mouse facilities and in consequence the microbiome represents a major source of uncontrolled variation, with the potential to significantly impact key experimental outcomes. This project, led by Jethro Johnson, a member of the Microbiome Cluster and PI at the Kennedy Institute of Rheumatology, will take advantage of the Network to systematically assess the extent of laboratory mouse microbiome variation across the UK.
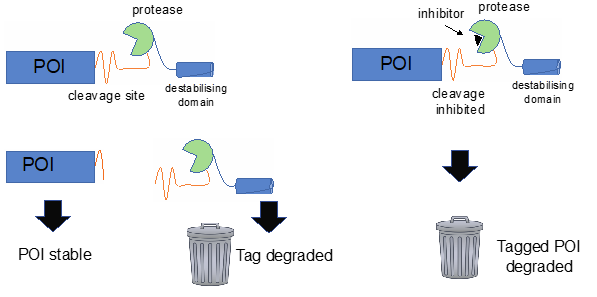
Another project with a direct relationship to a single cluster, but with broad application, is SMASh (Small Molecule Assisted Shut-off system), led by Degron Tagging Cluster lead Andrew Wood. The cluster makes use of tags that induce physical proximity to E3 ubiquitin ligases in order to direct proteasomal degradation. However, there are many proteins for which other methods will be required, such as proteins that localise within mitochondria or on the cell surface. This project will investigate an alternative approach for the inducible degradation of target proteins with a focus on proteins that are of interest to other Network partners, but for which inducible degradation is more challenging.
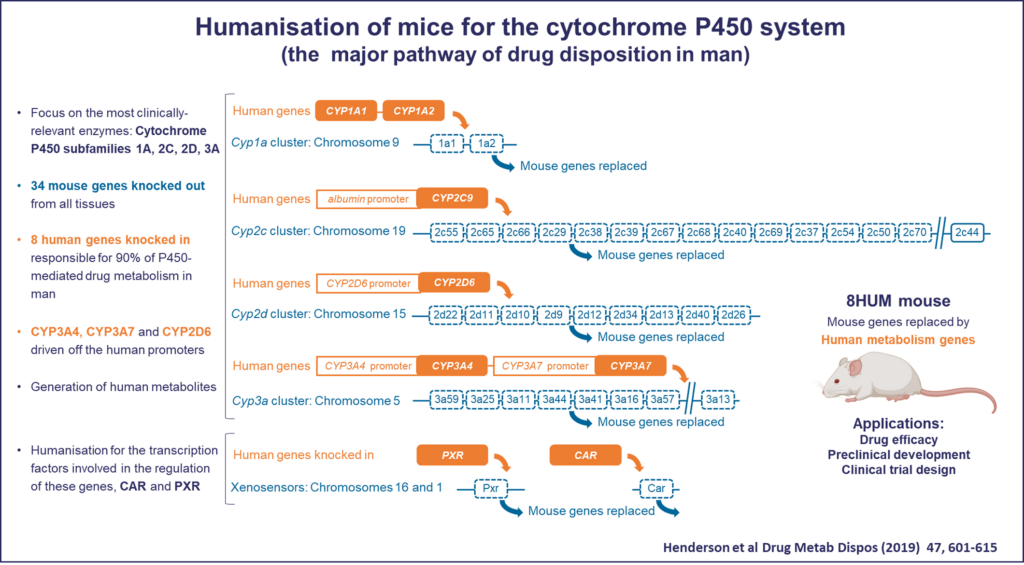
The project Evaluation of a humanised P450 mouse model for anticancer drug development involves a collaboration between members of the Cancer and Degron Tagging Clusters. It is led by Karen Blyth, lead of the Cancer Cluster, and Roland Wolf, member of the Degron Tagging Cluster, alongside Kevin Read, Head of Drug Metabolism and Pharmacokinetics at the Drug Discovery Unit at the University of Dundee. This project is addressing the challenge that there exist major differences between mouse and human drug metabolism, which is a significant contributing factor to why data obtained in mice does not extrapolate to the clinic. The team will test the utility of the 8HUM mouse, a line that has been humanised for the major genes involved in drug metabolism in man, in the development of anti-cancer drugs.
In contrast to the projects described above, although the project Understanding the function of a newly described mitochondrial protein in whole animal physiology is closely tied to a specific cluster, it is led by a Network Associate, Sally Clayton of the University of Birmingham, rather than a member of the Mitochondria Cluster. This project aims to understand the role of the AA467197 gene, which was recently shown to encode a small mitochondrial protein that localises to complex IV of the electron transport chain, but for which a precise function is not understood.
Similarly, Network Associates Antonella Spinazzola and Ian Holt, both at UCL, lead the project Detecting cholesterol in biological membranes in vivo: a new tool for biomedicine. Excess cholesterol is a major contributor to many human disorders, especially cardiac and neurodegenerative diseases, yet relatively few studies examine cholesterol’s behaviour in biological membranes in vivo. The project leads also recently identified an unexpected link between mitochondria and cholesterol homeostasis in a genetic form of mitochondrial disorder. In this project, they aim to create a mouse model that contains a reporter for cholesterol in cellular membranes and that could be used for the study of the full panoply of diseases characterised by aberrant cholesterol metabolism.

Finally, Sara Wells, Director of the Mary Lyon Centre, Dominique Bonnet from the Francis Crick Institute, and William Grey from the University of York, are establishing the UK patient-derived xenograft biobank. This collaboration aims to generate diverse, robust, and well-annotated patient-derived xenograft libraries across disease types. Projects undertaken will range from studying multiple blood cancers and testing drug efficacy, to providing humanised patient-derived xenografts to study and test potential immunotherapy responses in solid tumours.
Further details about all currently funded projects can be found on the Projects page. The next funding call from the Network Director’s Fund will open in early 2024. To find out more when the call opens, we recommend signing up to the National Mouse Genetics Network Newsletter.