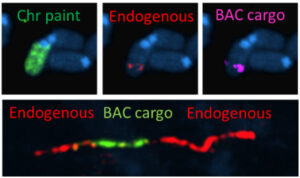
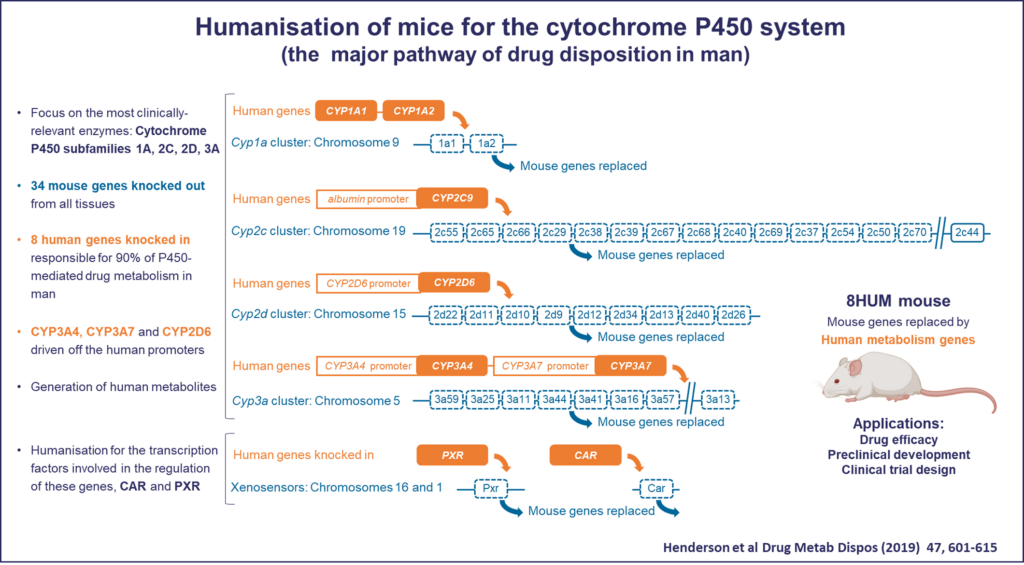
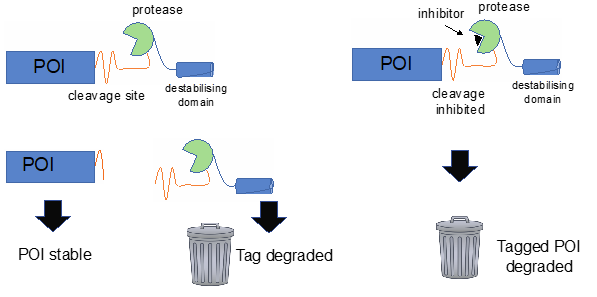
Although mutations and variation that contribute to human disease can be modelled in mouse, as we learn more about disease mechanisms, it is becoming increasingly clear that simple genetically altered mouse models may be of limited use for investigating human pathologies.
We know that genetic variation and mutation that is associated with disease risk often lies within regions of the genome which do not encode proteins. Yet there are limited technologies which allow these non-coding variations to be modelled in vivo.
Similarly, we are learning that the structure of genes plays an important role in their regulation and are identifying human diseases in which structural changes play an important role. Understanding gene regulation at scale necessitates large-scale gene engineering, technologies for which are frequently not routinely available within core facilities.
Establishing simple protocols to allow large regions of the mouse genome to be manipulated would unlock the potential of the mouse for modelling and interrogating all human genetic variation.
- Replacing large mouse sequences with the equivalent human sequences and building in the exact disease associated mutation could increase the accuracy of mouse models of human disease.
- Altering the structural elements of chromosomal DNA at scale would allow a better understanding of gene regulation and provide insights into perturbations that contribute to human disease.
How we do it
Working mainly in mouse embryonic stem cells, we will test, compare and optimize technologies for large scale engineering of genetic loci. Technologies which will be investigated include
- CRISPR/Cas9 associated gene targeting using Bacterial Artificial Chromosomes
- Recombinase Mediated Cassette Exchange
- Serine Recombinase (Integrase) mediated manipulations
In addition, under the remit of horizon scanning, we aim to implement and test newly described technologies to establish whether they provide a robust and reproducible methodology for disease modelling.
Once established and optimized, we hope to deliver a dedicated tool kit of selection cassettes, vector backbones and defined genetic strategies to help disseminate the streamlined protocols to the community.
Working with the network’s research theme clusters, we hope to develop proof-of-concept models which use the developed technology to model complex human disease in the mouse.