MitoCluster: an integrated phenotyping and mouse model generation platform for mitochondrial disease and dysfunction
Aims
Mitochondria are tiny energy generators that exist in large numbers (100s–1000s) inside human cells. Primary mitochondrial diseases (PMDs) are a large group of genetic disorders that impair mitochondria and the body’s ability to make energy. They can cause disabling symptoms that may impact very severely on quality of life. Though individually rare, as a group PMDs are a common cause of nervous system diseases. The effects of faulty mitochondria seen in PMDs are also present in other common diseases, such as dementia, diabetes and cancer. A better understanding of PMDs could also help us to understand these more common conditions.
PMDs are caused by alterations in DNA inside the cell nucleus or in the small amount of DNA (mtDNA) inside the mitochondria themselves. PMDs can cause symptoms at birth, in childhood or during adulthood, and cause a wide variety of medical problems, depending on the body parts most severely affected. As mitochondria are present in almost all cells, any system in the body can be affected. Muscles are particularly susceptible to reduced energy, which causes fatigue and weakness. There are currently no effective treatments for PMDs.
A barrier to PMD research, and that of other diseases in which mitochondria play a role, is a lack of relevant mouse models. Recreating PMDs in mice will allow researchers to confirm that genetic mutations cause PMD symptoms, find new ways to measure the disease over time and develop and test new treatments that can then more safely progress to trials in humans.
It has been very challenging to create mtDNA models of human PMDs in mice, but new tools to change mtDNA have recently been established. Also, modern techniques to measure mouse development, movement and behaviour that can be applied over long periods without stressing mice will allow researchers to study PMDs in these animals more accurately and humanely.
MitoCluster is a network of researchers based in the UK and Italy who aim to: (1) deliver new insights, therapies and biomarkers of PMDs; and (2) help understand the role of mitochondria in other common diseases (e.g., dementia, diabetes and cancer).
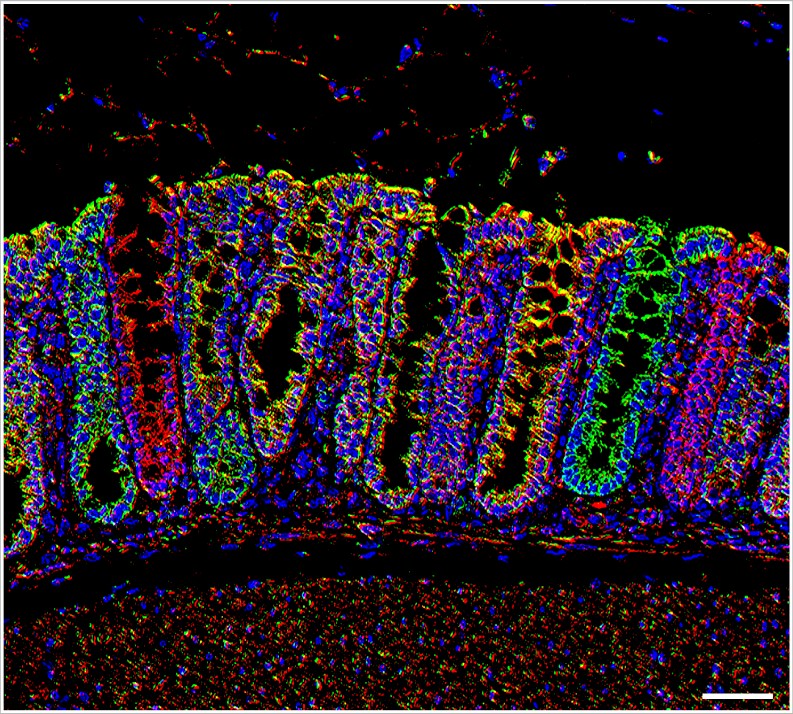
How we do it
Primary mitochondrial diseases (PMDs) caused by mutations in nuclear (nDNA) or mitochondrial DNA (mtDNA) that impair energy production, affect 1/4,300 in the UK. Despite causing severe disability and shortened lifespan, management is primarily aimed at ameliorating symptoms . This reflects an incomplete understanding of underlying pathogenesis and the fact that there are few reliable, non-invasive biomarkers to diagnose and monitor disease progression. The UK is at the forefront of PMD clinical and scientific research. MitoCluster is well-positioned to connect this world-leading expertise with mtDNA genome editing (mtGE) and deep phenotyping skills, to create a multi-site UK-wide platform for much-needed PMD mouse model generation, breeding and phenotyping.
Aim 1 (WP1) will generate a refined phenotyping platform, including measures of muscle fatigue and weakness, the leading targets of drug development given impacts on quality of life. We will combine in-cage assessments at Mary Lyon Centre with biochemical, imaging, metabolomics and single-cell sequencing at partner sites.
Aim 2 (WP2 and 3) will generate 6–9 novel mtGE mouse models using two approaches: (1) phenotype-first, in which breeding of heterozygous mitochondrial mutator (PolgA+/D257A) mice results in random heritable heteroplasmic mtDNA mutations — lines are selected via measurable phenotypes; (2) targeted, based on programmable nuclease (mtZFN) or cytosine base editing (DdCBE) methods to obtain predefined deleterious mtDNA mutations. These will be subject to deep phenotyping through our refined platform (WP1) and will undergo combined single-cell mtDNA/RNA-Seq in various tissues over time, to generate a publicly available atlas of somatic heteroplasmy dynamics.
We will harness our strong Pharma partnerships to evaluate the impact of existing and novel compounds in a whole-organism context (Aim 3) and develop and deliver courses in mtGE model generation and maintenance (Aim 4), to grow the UK skills base across academia and industry.
Cluster Members
| Name | Organisation | Contact |
|---|---|---|
| Dr Robert Pitceathly | UCL Queen Square Institute of Neurology | r.pitceathly@ucl.ac.uk |
| Prof Patrick Chinnery | MRC Mitochondrial Biology Unit / University of Cambridge | ClinNeuroHoDSec@medschl.cam.ac.uk |
| Dr Laura Greaves | Wellcome Centre for Mitochondrial Research / Newcastle University | laura.greaves@ncl.ac.uk |
| Prof Michael Hanna | UCL Queen Square Institute of Neurology | m.hanna@ucl.ac.uk |
| Dr Thomas Keane | EMBL-EBI | tk2@ebi.ac.uk |
| Prof Robert McFarland | Wellcome Centre for Mitochondrial Research / Newcastle University | robert.mcfarland@ncl.ac.uk |
| Dr Michal Minczuk | MRC Mitochondrial Biology Unit / University of Cambridge | mam@mrc-mbu.cam.ac.uk |
| Dr James Stewart | Wellcome Centre for Mitochondrial Research / Newcastle University | jim.stewart@ncl.ac.uk |
| Dr Jelle van den Ameele | MRC Mitochondrial Biology Unit / University of Cambridge | jv361@mrc-mbu.cam.ac.uk |
| Dr Carlo Viscomi | University of Padua | carlo.viscomi@unipd.it |
| Dr Alex Whitworth | MRC Mitochondrial Biology Unit / University of Cambridge | a.whitworth@mrc-mbu.cam.ac.uk |
Member Organisations

UCL Queen Square Institute of Neurology

MRC Mitochondrial Biology Unit
University of Cambridge

Wellcome Centre for Mitochondrial Research

Newcastle University
EMBL-EBI

