From Molecules to Models: Advancing Bowel Cancer Research through Collaboration and Mentorship

by Dr. Sudhir B. Malla, Senior Postdoctoral Researcher, MRC Mouse Genetics Network
As a cancer researcher within the MRC National Mouse Genetics Network (NMGN), my journey from a junior postdoctoral researcher to a senior author has been profoundly shaped by a collaborative and supportive environment. Operating across Belfast and Glasgow in Dr. Philip Dunne’s Lab, our team is dedicated to unravelling the complexities of bowel cancer – a prevalent and lethal disease in the UK.
Our recent endeavours, bolstered by the NMGN, have led to significant advancements in understanding colorectal cancer (CRC) biology and refining preclinical models to enhance treatment strategies.
Aligning Mouse Models with Human Colorectal Cancer Subtypes
In our study published in the British Journal of Cancer (Amirkhah et al., BJC, 2023), we addressed the challenge of aligning genetically engineered mouse models with the consensus molecular subtypes (CMS1-4) of human CRC. By developing the MmCMS classifier, we provided a robust tool to match mouse models with human cancer subtypes, facilitating more accurate preclinical testing and translational research.
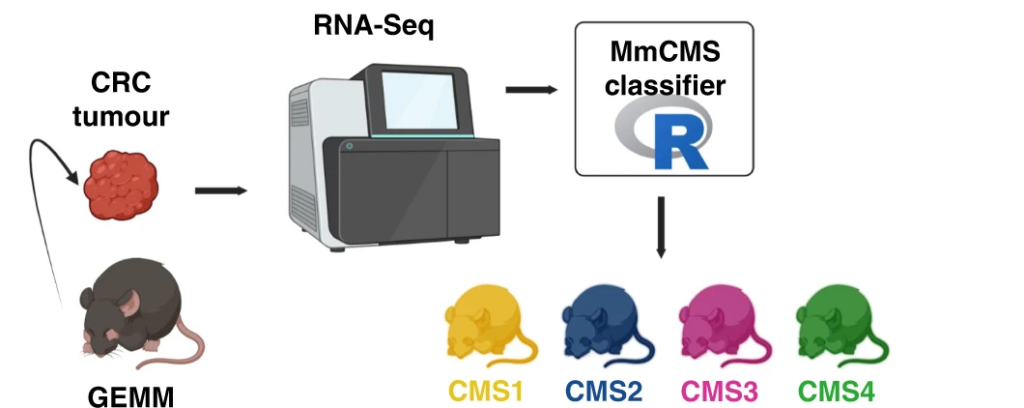
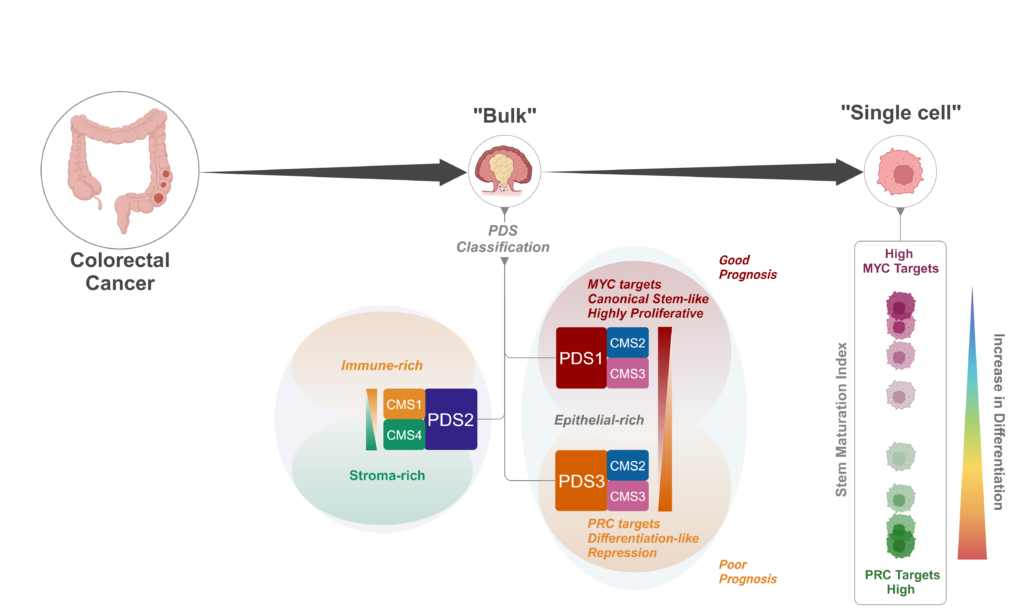
Discovering a Slow-Cycling Tumour Phenotype with Poor Prognosis
Our research featured in Nature Genetics (Malla et al., Nature Genetics, 2024) introduced pathway-derived subtypes (PDS) to classify colorectal tumours based on biological pathways rather than individual gene expression. Notably, we identified PDS3 – a slow-cycling tumour phenotype within CMS2/3, characterised by reduced stem cell populations and increased differentiation. Despite its less aggressive appearance, PDS3 is associated with poor clinical outcomes, highlighting the need for tailored therapeutic approaches.
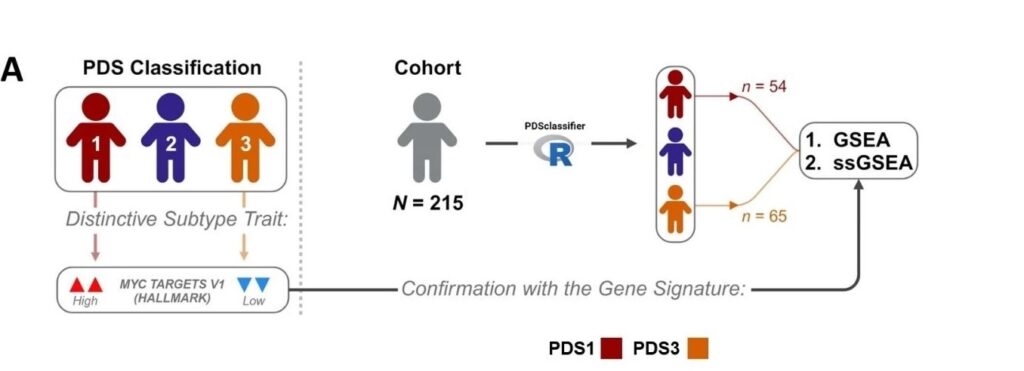
Integrating Computational Tools for Enhanced Biological Discovery
Building upon these findings, we developed dualGSEA, an R-based computational tool detailed in Scientific Reports (Bull et al., Scientific Reports, 2024). This tool enables simultaneous statistical and biological analysis of gene expression data to explore important pathways in comparative studies for their significance and predictability. By facilitating more nuanced interpretations of transcriptomic data, dualGSEA aids in identifying potential signatures and biomarkers for therapeutic targets.
The Impact of Mentorship and Collaborative Networks
The NMGN has been instrumental in my professional development, offering mentorship in both cancer biology and computational research. The Network’s collaborative framework has provided access to diverse expertise and resources, fostering an environment conducive to scientific innovation and career progression.
“We often talk about outputs, but the development of people is just as important. Supporting early-career researchers like Sudhir is essential to the future of UK science.”
– Dr. Philip Dunne, Principal Investigator
“Sudhir’s journey and his research achievements exemplify what the MRC National Mouse Genetics Network is here to do – supporting high-quality science and fostering the next generation of research leaders.”
– Professor Owen Sansom, Director of the CRUK Scotland Institute
Looking Ahead
Through the integration of molecular biology and computational tools, our research continues to advance the understanding of colorectal cancer. The support and mentorship provided by the NMGN not only enhance scientific discovery but also cultivate the growth of emerging researchers, ensuring a robust future for cancer research.
With the recent launch of the CRC-STARS project – a major international research initiative jointly funded by Cancer Research UK (CRUK), the Bowelbabe Fund for Cancer Research UK, philanthropic support from Bjorn Saven CBE and Inger Saven, and the Scientific Foundation of the Spanish Association Against Cancer (FCAECC), I have been appointed as a Co-investigator and a Computational Lead for the Early Career Researcher (ECR) Steering group. In this role, I represent a network of computational and mathematical ECRs across the consortium, fostering collaboration, promoting interdisciplinary integration, and facilitating knowledge exchange.