A new tool for rapid protein degradation in live animals
A primary way in which we have developed our understanding of many diseases is via control of gene function, whether observed in cells grown in a dish or in live animal models. This has generally been done by manipulating DNA or RNA, but new research from Andrew Wood, lead of the MRC National Mouse Genetics Network’s Degron Tagging cluster and Group Leader at the MRC Human Genetics Unit at The University of Edinburgh, has provided a proof-of-principle for a new way of studying protein function in mouse models of development and disease.
Throughout the history of genetics and molecular biology, scientists have sought to understand disease by disrupting normal physiology by deleting, mutating, or silencing genes or by identifying genetic variants in disease states. But biological processes are largely driven by proteins, so new methods that directly manipulate proteins could be of great benefit. One major advantage of this is that methods that drive degradation of a target protein will work much faster than those that prevent production of new protein by silencing expression of the target gene.
Additionally, although modern transgenic technologies enable the deletion of genes in a time- and location-controlled manner, these changes are hard to reverse, so a method that can remove protein and then restore it to normal levels could be of great benefit to our understanding of the roles of proteins at different stages. Therefore, techniques that drive degradation of proteins should aim to be rapid, reversible, and targeted.
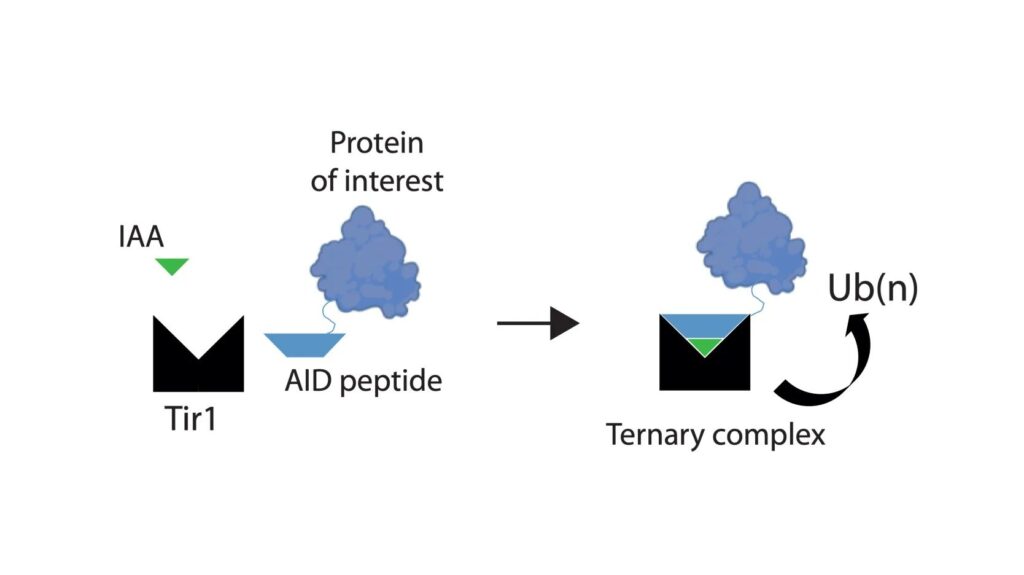
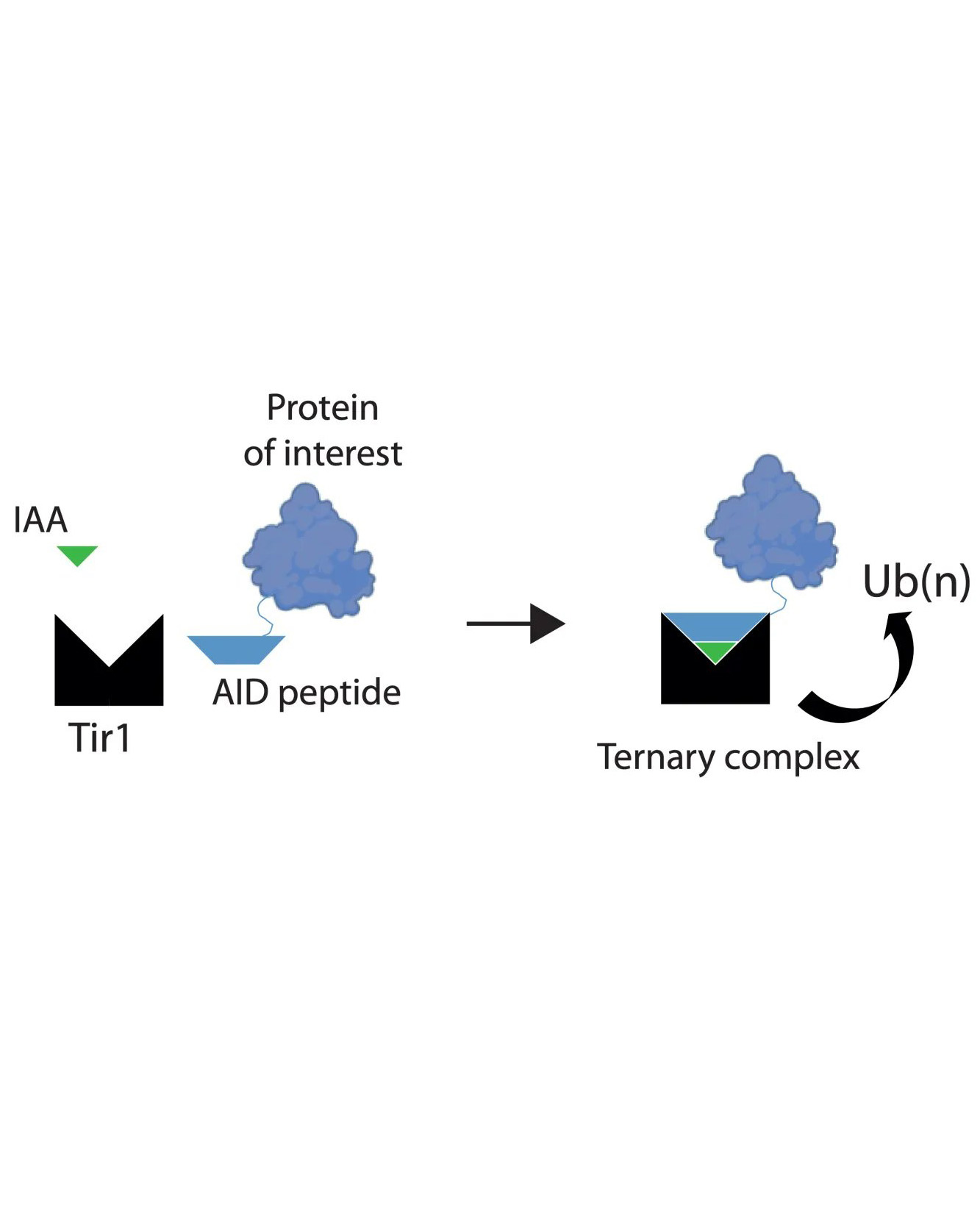
Auxin inducible degrons are a chemical genetic tool that might provide this solution. Auxins are plant hormones that bind to Tir1, a subunit of a plant ubiquitin ligase complex, and increase the affinity of Tir1 for proteins that contain a degron polypeptide. This formation of a larger complex allows ubiquitination of the degron-tagged protein, a system cells use to mark a protein for degradation by the proteasome. Development of this system in non-plant cells has revolutionised functional studies of proteins in mammalian in vitro systems, but it has not yet been possible to degrade tagged endogenous proteins in adult tissues of living animals. Such an ability would benefit the study of a wide range of models of normal physiology and disease.
In their new paper, Andrew’s group have derived novel transgenic mouse lines that use this degron system to investigate the functions of two protein subunits of condensin complexes that are essential for chromosome formation and segregation during mitotic cell division. They were able to achieve near complete (>90%) protein degradation within 2 hours. Although degradation was seen in most cell-types tested, issues such as absent components of the ubiquitin proteasome system or poor transit of auxin across barriers within tissues meant that degradation was not possible in a minority of cell-types.
The team also found that the dosage of the target protein, auxin ligand, and Tir1 can all determine the kinetics of protein degradation. One important effect of this is that it might make highly expressed proteins more challenging targets for chemical degradation strategies. Similarly, in the absence of auxin ligand, there was no degradation, dispelling previous concerns that Tir1 alone might be able to bring the ubiquitin ligase and protein target together to drive proteasomal degradation.
With respect to their investigation of subunits of condensin I and condensin II, they found that both complexes are needed for cell division in precursor lymphocytes, but not in differentiated peripheral lymphocyte derivatives, where cells can divide with only one complex or the other. Rapid removal of proteins is key to studying essential cell cycle proteins such as condensins, as secondary or tertiary phenotypes arising from abnormal cell division can quickly obscure the primary effects of loss of the degraded protein.
This study is an important first step for our Degron Tagging cluster, the aim of which is to develop protocols for protein degradation across tissues, diseases and timescales in a way that can reduce and refine animal use. As most drugs target proteins, these tools could also provide a useful way to model the effects of drugs and other therapeutic interventions, alongside their use for functional studies of proteins in disease models. Another core aim of the cluster is to share best-practice in these technologies via training courses and an online information portal, so that they can best complement, extend, or replace existing approaches and the development of robust and reliable technologies will form a strong basis for this aim.
This study was performed in collaboration with Dr Bin Gu (Michigan State University) and Professor Janet Rossant (Sick Kids, Toronto) and supported by UKRI MRC, Wellcome Trust, and Canadian Institutes of Health Research.
Macdonald, L, Taylor, GC, Brisbane, JM, Christodoulou, E, Scott, L, von Kriegsheim, A, Rossant, J, Gu, B, Wood, AJ. Rapid and specific degradation of endogenous proteins in mouse models using auxin-inducible degrons. eLife (2022) 11:e77987 https://doi.org/10.7554/eLife.77987