Understanding the Protein Tagging Problem: A New Study from the NMGN Degron Tagging Cluster

Celebrating Research Specialists: Driving Innovation in Mouse Genetics
Research specialists are often the unsung heroes in biomedical research groups, providing continuity in long-term research programmes, building up a wealth of expertise across diverse techniques, and training junior researchers whilst leading their own innovative projects. Dr Gillian Taylor exemplifies this vital role as a postdoctoral research specialist in the National Mouse Genetics Network’s Degron Tagging Cluster, where she has been instrumental in translating cutting-edge protein control technologies into mouse models.
In a 2022 eLife paper, Gillian and colleagues demonstrated that mouse proteins engineered with degron tags could be selectively degraded in living tissues using chemical triggers. This technology holds promise for validating drug targets in vivo and laid the foundation for the current study.
The Protein Tagging Problem: A Barrier to Genetic Innovation
One of the major challenges in applying degrons and other genetic technologies in mice is the protein tagging problem; the risk that adding tags to proteins can alter their function in unpredictable ways. This has serious implications for the 3Rs principles, as poorly designed models may lead to unnecessary animal use or misleading results.
While the tagged constructs used in the eLife study performed well in cell lines, their behaviour in mouse models was unexpectedly variable. The new PLoS Genetics paper explores these tissue-specific effects in detail, revealing that the same tag can stabilise a protein in one tissue while promoting its degradation in another.
Dr Taylor’s paper details these surprising findings, demonstrating how the controlled laboratory environment of cell lines can mask effects that become apparent in the complex physiological context of living mice.
Key Findings: Tissue-Specific Effects of Tag Fusions
Using genome-edited mouse models, the team uncovered two distinct mechanisms by which protein tags can affect expression:
- Degradation Leakage: The auxin-inducible degron (AID) is one of the most widely use degron tags. In tissues like the small intestine, high levels of the inducer molecule indole-3 acetic acid (IAA) produced by the microbiome triggered unintended degradation of AID-tagged proteins. This effect was dependent on the presence of the transport inhibitor response 1 (TIR1) receptor and could be mitigated using the AID2 system, which is less sensitive to IAA.
- Degron Occlusion: Tags are typically fused to the N- or C-terminus of a protein. However, protein termini often contain natural degron motifs that promote protein destruction in certain contexts. Fusion of tags at these termini prevents destruction, leading to increased protein stability. This effect was particularly evident in post-mitotic cells and could be reversed by duplicating the terminal amino acid sequence during allele design.
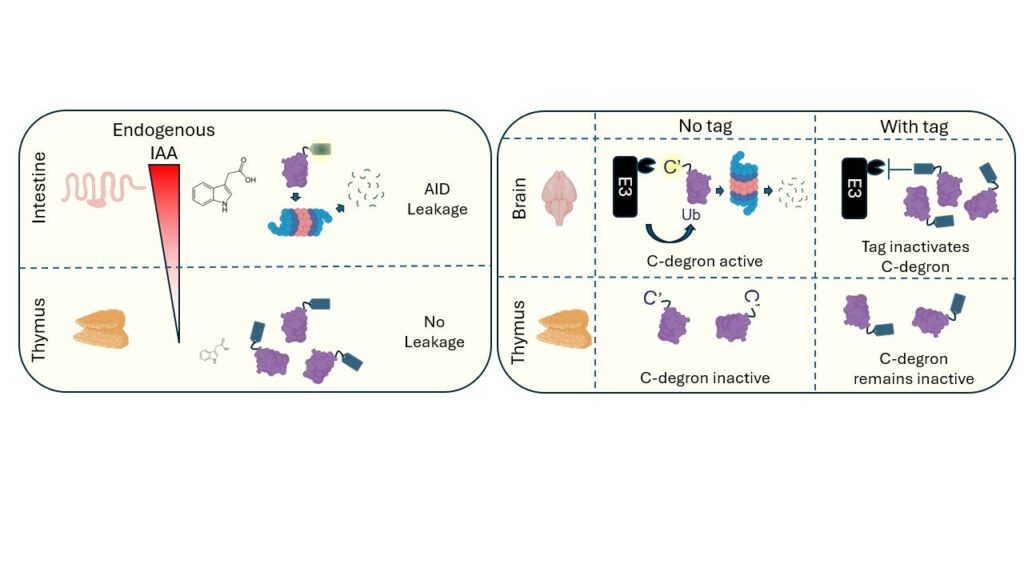
Left. AID leakage. In the intestine, high levels of the inducer molecule IAA are produced by the microbiome, triggering leaky TIR1-dependent destabilisation of AID-tagged NCAPH and NCAPH2 proteins. The AID2 system uses a different chemical inducer and is 7-fold less sensitive to IAA-mediated leakage. Right: Degron occlusion: N- and C-degrons at endogenous protein termini can be active only in specific tissues (e.g. brain). For NCAPH2 and SOX2, Tag fusion inactivates degron function, which only affects protein levels in tissues where the degron was previously active. Degron occlusion can be rescued by reinstating terminal amino acids.
Broader Implications for Mouse Genetics
These findings underscore the need to understand normal mechanisms governing protein stability to enable protein engineering strategies that leave normal regulation intact. Resources such as Degronopedia, a web-based tool that predicts natural degrons, can help steer researchers away from fusion sites that disrupt natural degrons.
This, in turn, has wide-reaching implications for allele design, model validation, and genetic engineering best practices. It reinforces the value of collaborative, cross-disciplinary approaches championed by the NMGN and provides practical strategies for improving the reliability of tagged protein models. It also underscores how the Network’s support structure enables research specialists to make substantial contributions to the field and drive innovation, whilst advancing their own professional development.
“Working as part of the NMGN has provided great opportunities for me to grow as a researcher by further developing my skills base through training courses and providing new opportunities for collaboration across the UK. We work closely with experts in disease modelling, which makes it easier to translate our work on genetic technologies into real-world applications.”
— Dr Gillian Taylor, lead author
“Research specialists and technical staff play a vital role in biomedical research labs across the UK. Gillian has been at the centre of our work in the NMGN cluster, and it is great to see her work recognised in this first author publication.”
— Dr Andrew Wood, senior author and Degron Tagging Cluster lead