Precision control of protein dosage in vivo
Aims
Technologies to change gene function underpin mouse genetics. However, mouse genes are normally disrupted by mutating DNA, whereas most biological functions are carried out by proteins. This has several drawbacks, which currently limit the ability of mice to model human disease mechanisms and therapeutic interventions:
- Timescale: After DNA is mutated, protein function is lost gradually, sometimes over several days. This gives cells and tissues time to adapt and makes it difficult to identify direct protein functions and understand disease mechanisms.
- Reversibility: DNA changes are hard to reverse. Better ways to toggle between protein “on” and “off” states in living mice would enhance our ability to determine whether, and at what critical time-points during disease progression, therapeutic interventions could yield clinical benefit.
- Dosage control: Many human disease states involve incomplete reductions in protein dose, which are often poorly modelled by mutating DNA. The ability to titrate the dose of proteins at different stages of the disease process is therefore highly desirable.
- Modelling drug activity: Many drugs target proteins rather than nucleic acids. To convert more preclinical leads into effective therapies, we need better methods to validate potential drug targets in human disease models.
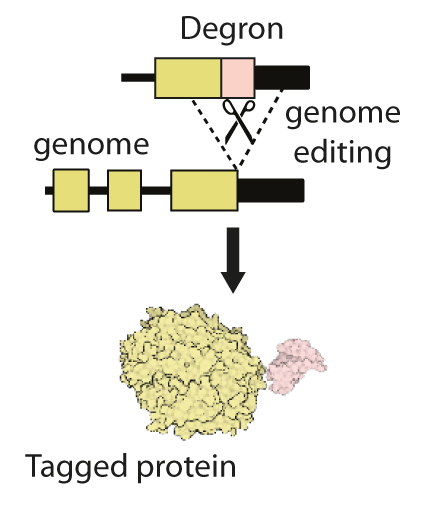
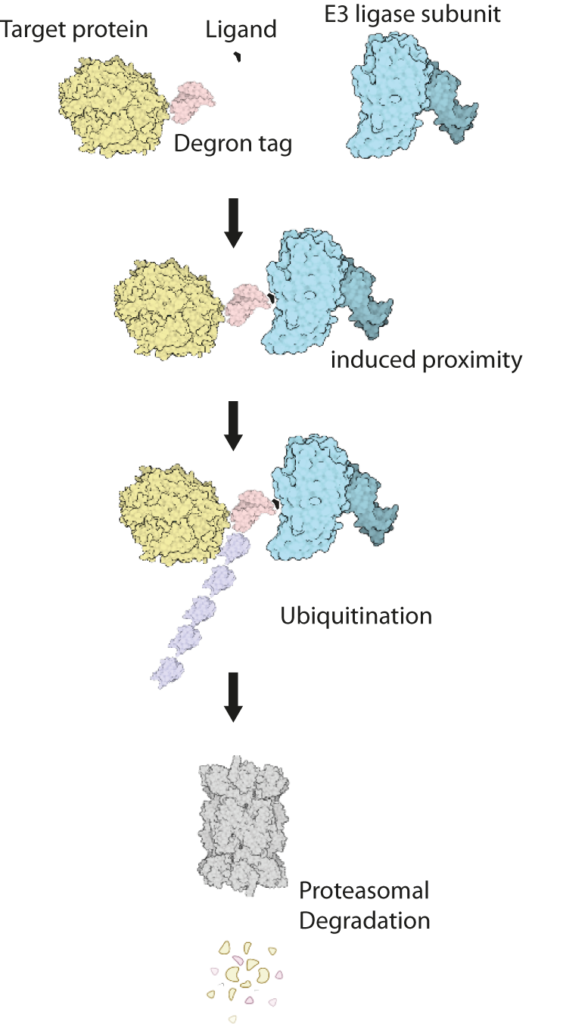
This cluster will combine recent technological developments in the engineering of genomes, proteins and small molecules to address these unmet needs. We will use existing gene editing approaches to add short ‘tags’, known as ‘degrons’ onto proteins of interest (Figure 1). Degron-tagged proteins can then be targeted by small drug-like molecules that can enter cells and hijack their natural protein-destruction pathways to facilitate rapid, tuneable and reversible degradation (Figure 2).
Degron tags are already widely and successfully used in cultured cells and invertebrates. However, research on their use in mouse models is at an early stage, and the complex environment of mammalian tissues poses several new challenges. To address these challenges, our cluster team brings together expertise in genome editing, protein engineering, protein turnover, medicinal chemistry and pharmacology. The MRC National Mouse Genetics Network will facilitate interactions with leading disease modellers to translate degron technologies into mouse models of human disease.
How we do it
The workplan will proceed in two phases. During phase I, genetic reporters will be built to evaluate and compare different degron systems across tissues and disease models. We will test existing ligands and synthesise novel derivatives to identify compounds with favourable pharmacokinetic and pharmacodynamic properties in vivo. Our aim is to develop protocols for protein degradation across tissues, diseases and timescales that are in line with the ‘3R’ principles and network priorities. During phase II, we will work with the Mary Lyon Centre to establish a national hub for degron technologies, disseminating knowledge from phase I across the network via exemplar projects, and to the wider community via training courses and an online information portal. Our vision is that these tools will complement, extend, and in many cases replace recombinase-based approaches, simultaneously reducing and refining animal use while producing data that are easier to interpret.
Cluster Members
| Name | Organisation | Contact |
|---|---|---|
| Dr Andrew Wood | MRC Human Genetics Unit / University of Edinburgh | andrew.j.wood@ed.ac.uk |
| Dr Joe Marsh | MRC Human Genetics Unit / University of Edinburgh | joseph.marsh@ed.ac.uk |
| Prof Gopal Sapkota | MRC Protein Phosphorylation Unit / University of Dundee | g.sapkota@dundee.ac.uk |
| Prof Asier Unciti-Broceta | CRUK Edinburgh Centre / University of Edinburgh | asier.ub@ed.ac.uk |
| Prof Roland Wolf | University of Dundee | c.r.wolf@dundee.ac.uk |
Member Organisations

MRC Human Genetics Unit

University of Edinburgh

MRC Protein Phosphorylation and Ubiquitylation Unit

University of Dundee

