Patient-led functional genomics
Aims
Approximately 1 in 20 babies are born with severe anatomical malformations. Each year this equates to 8 million affected newborns, of which 300,000 die within the first four weeks of life. With recent advances in sequencing technology, we are accelerating the identification of possible disease-causing changes in the genetic code of these patients. However, it is a major challenge to prove which of these genetic changes, also called variants, do cause these malformations, as well as to establish the cellular mechanisms by which these changes disrupt normal development.
How do we distinguish one problematic inherited or spontaneous variant in DNA from the many benign changes, and prove that it disrupts normal development? Can we better understand why some patients are more affected than others even though they carry similar, if not the same, genetic changes? How do important environmental influences, such as maternal health during pregnancy, modify how these genetic changes present themselves in terms of severity and spectrum of presentations observed in patients? Many of the genes implicated in congenital anomalies play multiple roles in different tissues during prenatal and postnatal development; thus, these genes are difficult to study in humans, even in stem cell ‘disease-in-a-dish’ models.
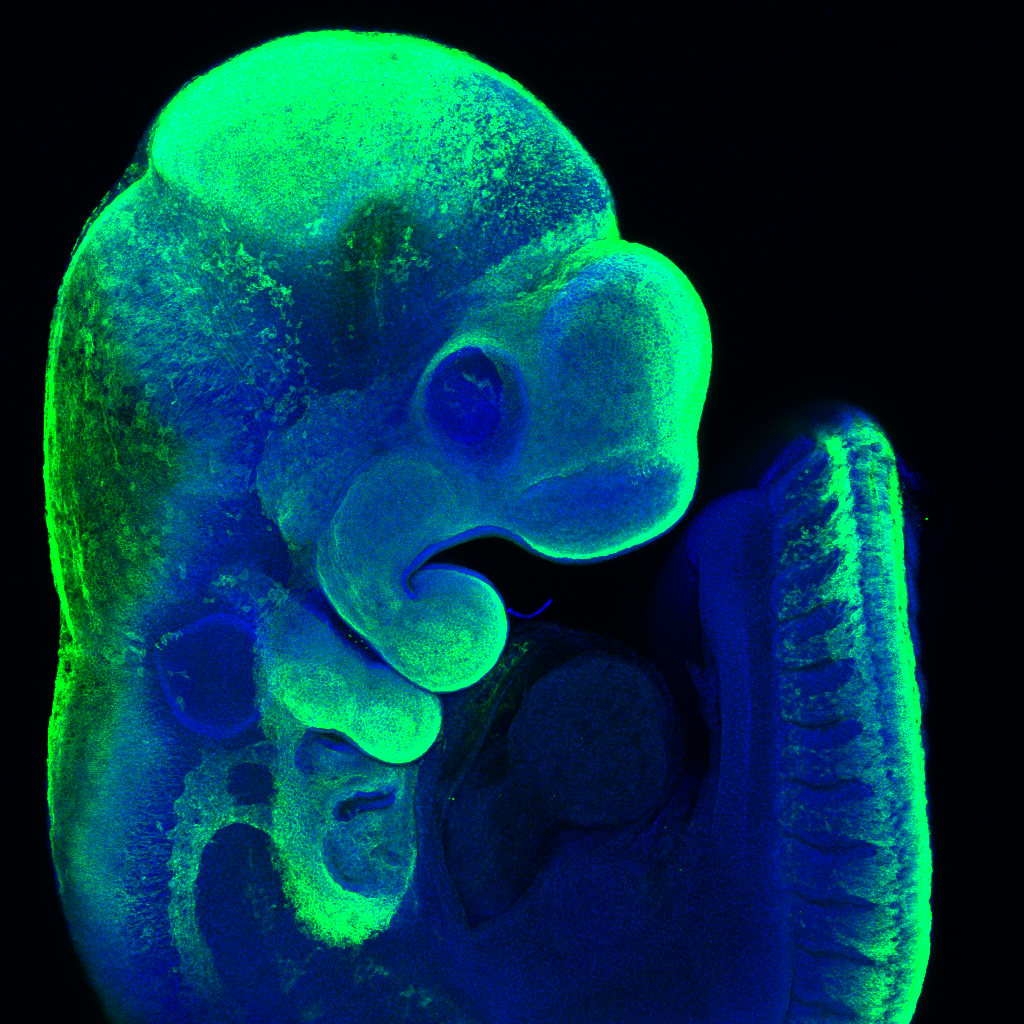
Our goal is to make precisely engineered mouse models of patient variants, which will help us to replicate complex interactions disrupted during early life, across multiple organ systems. We also aim to improve automated live monitoring of early life in our animal models, which will help us to better understand the consequences of these genetic mutations during the critical postnatal period. Novel mouse models will also allow us to monitor disease progression later in life and will serve as platforms for developing much needed therapeutic interventions.
Our integrated programme will improve the use of animal models, while advancing the basic research into early life anomalies. We will be able to improve our discussions on genetic cause and effect together with clinical geneticists, medical teams and their patient groups. The ultimate hope is to provide improved diagnoses and prognoses for patients with congenital anomalies.
How we do it
This cluster will generate and study mouse models of prioritized gene variants identified from patients with congenital anomalies, focusing on anomalies affecting the cranial, neural, heart and kidney structures. For more information and to submit a VUS please follow this link. We will use genome engineering to mimic the human gene variants in mouse models, in order to assess the overall functional consequence of pathogenic mutations. The cluster will analyse and distribute these mutants, determine underlying causes, and collaborate with clinicians. A key objective is bringing together diverse experts studying syndromic disorders, as many genetic disorders affect multiple organ systems. A second objective is to improve live monitoring of animals in early life, which will improve our ability to link gene variation to function in structural malformations. The overall goal is to enhance UK expertise in determining causes, understanding mechanisms and identifying potential therapies for congenital anomalies.
New VUS call now open!
For more information and to submit a VUS please follow this link. We will use genome engineering to mimic the human gene variants in mouse models, in order to assess the overall functional consequence of pathogenic mutations. The cluster will analyse and distribute these mutants, determine underlying causes, and collaborate with clinicians.
Cluster Members
| Name | Organisation | Contact |
|---|---|---|
| Prof Karen Liu | King’s College London | karen.liu@kcl.ac.uk |
| Prof Nicholas Greene | UCL Great Ormond Street Institute of Child Health | n.greene@ucl.ac.uk |
| Prof Deborah Henderson | Newcastle University | deborah.henderson@ncl.ac.uk |
| Prof Pleasantine Mill | MRC Human Genetics Unit / University of Edinburgh | pleasantine.mill@ed.ac.uk |
| Dr Stephen Twigg | MRC Weatherall Institute of Molecular Medicine / University of Oxford | stephen.twigg@imm.ox.ac.uk |
Member Organisations

King’s College London

UCL Great Ormond Street Institute of Child Health

Newcastle University

MRC Human Genetics Unit

University of Edinburgh

MRC Weatherall Institute of Molecular Medicine


