Novel tools for modelling normal and perturbed haematopoiesis
Aims
Blood cell production (haematopoiesis) is a finely tuned process, producing trillions of new blood cells daily. Dysregulation leads to a wide array of haematological diseases, including bone marrow failure and leukaemia. Despite recently generating detailed molecular profiles of individual blood-forming cells at unprecedented resolution and comprehensively mapping genetic drivers in numerous haematological diseases, we lack effective tools to map and manipulate key cellular subsets involved in normal and stressed haematopoiesis, which would provide fundamental insight into disease aetiology and potential intervention strategies.
Our cluster of world-leading experts in developmental and malignant haematopoiesis, the haematopoietic niche, and oligogenic mouse modelling will develop new tools to revolutionise how we study and manipulate haematopoietic/immune-cell function for clinical benefit. Building on our extensive clinical and molecular datasets from normal, post-infection and pre-leukaemic and leukaemic cells, we will develop new tools to precisely map and manipulate haematopoietic cells. These approaches will support fundamental breakthroughs in haematopoiesis and facilitate production of new models with genetic manipulations restricted to highly defined cell populations with temporal regulation. This collaborative network will also involve cross-institutional model sharing and standardisation, and catalyse disease model development in other fields.
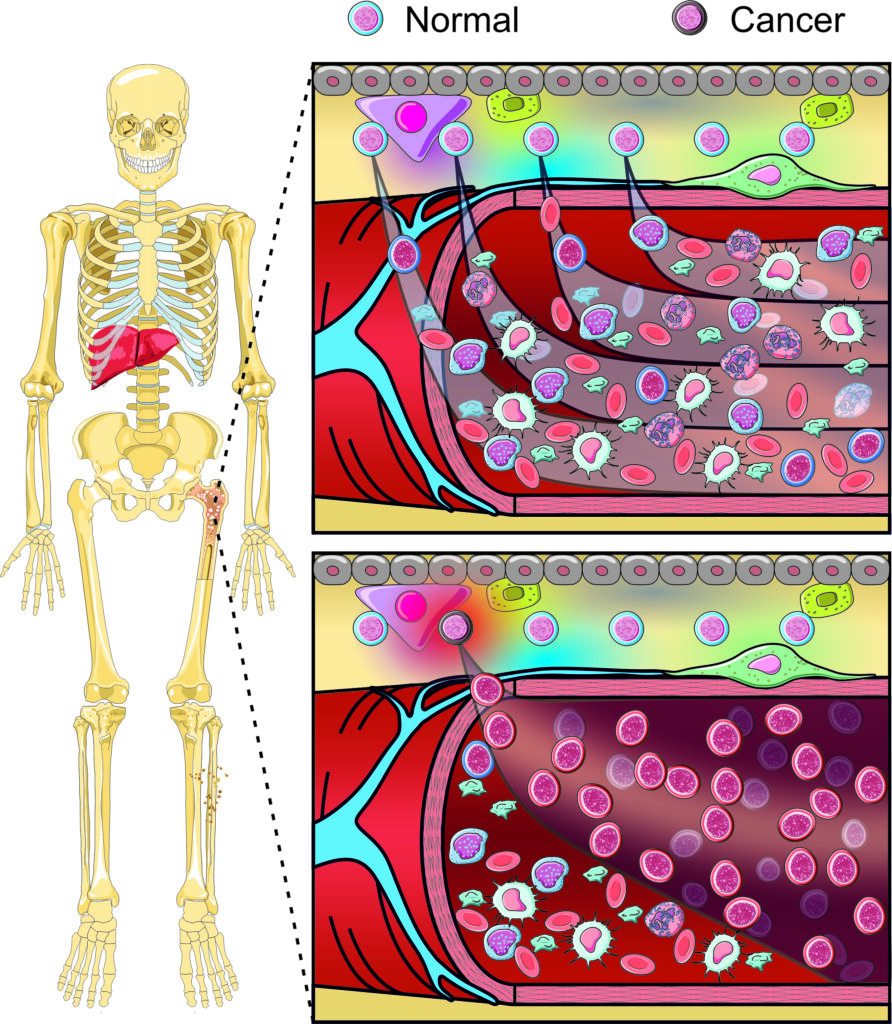
How we do it
Despite remarkable progress in the transcriptional and genomic profiling of normal and diseased blood cells, an urgent need exists to develop tools that will permit the dissection of the cellular and molecular mechanisms underpinning normal haematopoiesis and its responses to stress and disease. Tools to accurately map the cellular identity of specific haematopoietic cell subsets in vivo are regularly lacking and even fewer tools exist to selectively manipulate the expression of multiple genes in specific target cells in the correct spatio-temporal manner.
Building on our extensive transcriptomic and genomic datasets from normal, post-infection and pre-leukaemic cells in patients and mouse models, our cluster will work with the Mary Lyon Centre to develop next-generation mouse models, harnessing restricted intersectional recombinase gene expression to precisely map and manipulate haematopoietic cells. Specifically, our work will focus on three key areas: 1) new tools to mark and target unique haematopoietic cell types, 2) improved modelling of complex disease in haematological malignancies with respect to mutation timing and cell of origin and 3) new tools to specifically target normal and disease cells during development.
Cluster Members
| Name | Organisation | Contact |
|---|---|---|
| Dr David Kent | University of York | david.kent@york.ac.uk |
| Prof Brian Huntly | Wellcome-MRC Cambridge Stem Cell Institute / University of Cambridge | bjph2@cam.ac.uk |
| Prof Cristina Lo Celso | Imperial College London / Francis Crick Institute | c.lo-celso@imperial.ac.uk |
| Dr Andrew McKenzie | MRC Laboratory of Molecular Biology | anm@mrc-lmb.cam.ac.uk |
| Prof Claus Nerlov | MRC Molecular Haematology Unit / MRC Weatherall Institute of Molecular Medicine / University of Oxford | claus.nerlov@imm.ox.ac.uk |
| Prof Katrin Ottersbach | University of Edinburgh | katrin.ottersbach@ed.ac.uk |
Member Organisations

University of York

Wellcome-MRC Cambridge Stem Cell Institute
University of Cambridge

Imperial College London

Francis Crick Institute

MRC Laboratory of Molecular Biology

MRC Molecular Haematology Unit

MRC Weatherall Institute of Molecular Medicine

University of Oxford

